Self-Assembly of Proteins for New Materials
Designing the protein network.
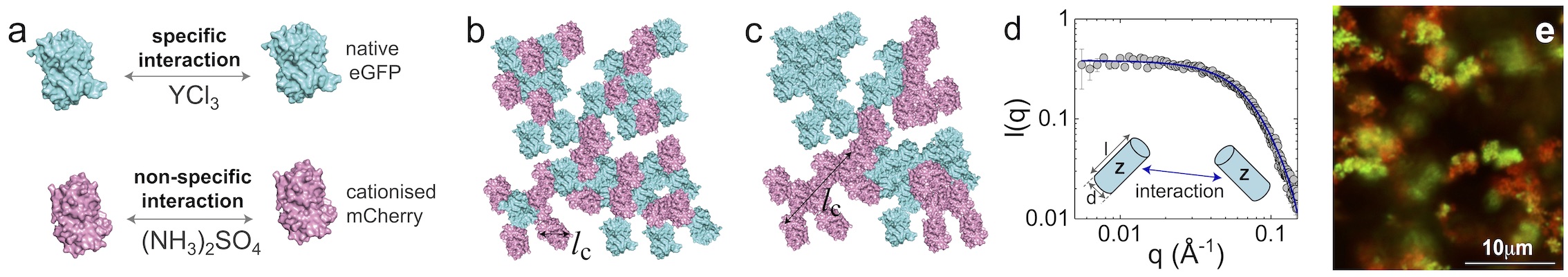
a-c: The self assembly stragety. d: The SAXS data. e: The confocal image.
The implementation of natural and artificial proteins with designer properties and functionalities offers unparalleled opportunity for functional nanoarchitectures formed through self-assembly. However, to exploit the opportunities offered we require the ability to control protein assembly into the desired architecture while avoiding denaturation and therefore retaining protein functionality. We address this challenge with a model system of fluorescent proteins. Using techniques of self-assembly manipulation inspired by soft matter where interactions between components are controlled to yield the desired structure. We have shown that it is possible to assemble networks of proteins of one species which we can decorate with another, whose coverage we can tune. Consequently, the interfaces between domains of each component can also be tuned, with applications for example in energy transfer. Our model system of fluorescent proteins eGFP and mCherry retain their fluorescence throughout the assembly process, thus demonstrating that functionality is preserved.